Undoubtedly, the one single technique that has revolutionized molecular biology was the introduction PCR (polymerase chain reaction). It has numerous applications in many different fields ranging from scientific and medical research to diagnostics, criminology, forensics, archeology and paleontology. In research PCR has several different uses as it has allowed things to be done that were unimaginable before, or just made things technically possible in a much shorter time. Kary Mullis was awarded the Nobel prize in Chemistry in 1993 for PCR improvements that allowed a fast and exponential amplification of pieces of DNA- he has been a bit controversial afterwards regarding his views on other subjects which you can read more about elsewhere on the web.
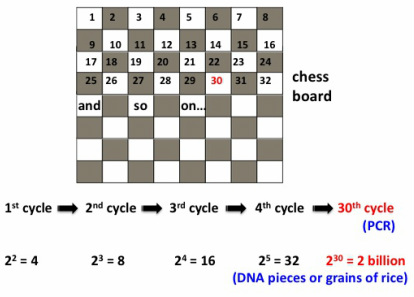
In essence, PCR is a technique that "amplifies" a piece of DNA, especially if the amount we have to begin with is so small that unless we get more, there is not much we can do with it. The DNA sample to be amplified from is called "template". To give an idea of how much DNA amplification you can get with PCR, I will use a story about the invention of chess and rice grains that I heard from my dad (who happens to be a mathematician) when I was a little girl. It goes like this: the king of the country where the inventor of chess lived (some versions say he commissioned the guy, who might have been a mathematician, to come up with a fun game) asked him what he'd like for his invention. The man asked the king to get one grain of rice for the first square of the chess board, two for the second one, four on the third one, and so forth, doubling the amount each time. The king, who thought this was a low price, ordered the treasurer to count the rice and give it to the inventor. The treasurer took more than a week to calculate the amount and explained that it would take more than all the rice in the kingdom to reward the inventor. The total number of grains covering the 64 squares of the chess board would be 18,446,744,073,709,551,615.
PCR amplification follows the same exponential growth: let's say we have one molecule (piece) of DNA in the tube to begin with, although usually there are more pieces in the initial sample. After the first PCR cycle we will have a total of 2 identical pieces of DNA after the action of the DNA polymerase enzyme (see details below). We now enter the second cycle, where the polymerase will make 2 more copies from the existing ones to result in a total of 4 .... and so on. Typical PCR reactions are setup with anywhere between 20-60 cycles.
By the 30th cycle there are already more than 1 billion copies per starting template molecule: as shown in the figure below, there will be exactly 2 billion pieces of DNA on the 30th cycle for each template DNA piece, or 2 billion grains of rice on the 30th chess board square.
https://www.youtube.com/watch?v=eEcy9k_KsDI
WHAT ARE THE INGREDIENTS AND HOW DOES THE REACTION WORK?
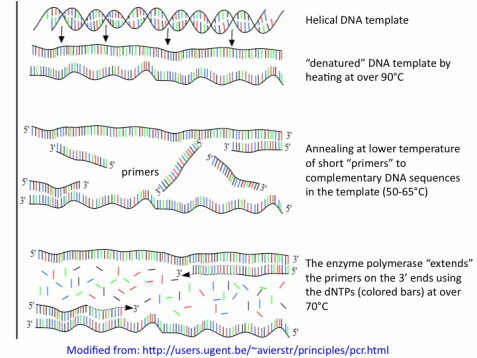
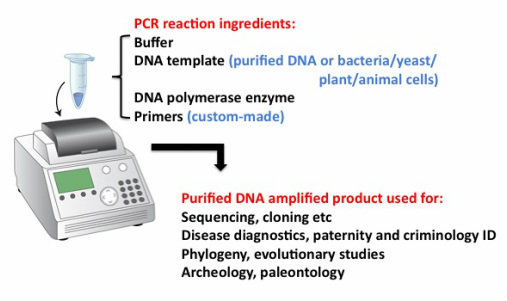
We might want to "sequence" a piece of DNA we isolated from an organism of interest or an animal or person with a disease of interest or from a crime scene or a fossil. Whatever the material is (human or animal tissue or cells (sometimes containing virus or bacteria as the targets we want to amplify), bone remains etc) the first step is to try to get purified DNA, for which there are commercial kits available that PCR laboratories buy routinely. Afterwards, a mix is set up in a tiny plastic tube or well plate containing the DNA and all the reagents necessary for the PCR reaction. Once again, most of these are purchased from specific companies and in this case consist of special buffers, DNA base units or deoxynucleotide triphosphates (dNTPs in 4 different flavors: G, A, C and T) and most importantly, the "enzyme" DNA polymerase which has made all this possible. The most widely used is a heat-stable form isolated from Thermus aquaticus (an organism that lives and functions at very high temperatures) and referred to for short as Taq.
By far the most important variable element of the PCR reaction is one that the researcher has to "design" (sometimes with the help of programs) to make the DNA amplification possible and specific for the target sequence. These are small pieces of synthetic DNA that one can order to be made based on a sequence provided and called "primers". There are usually 2 primers per piece of DNA to be amplified which flank the target sequence, each of them is complementary to one of the 2 strands of DNA in the sequence.
The PCR reaction tube is then placed into a PCR machine or cycler where one can run "programs" which will expose the contents in the tubes to specific cycles of different temperatures that will result in (see figure below): 1) denaturing of the double stranded DNA so that next the primers can 2) anneal to complementary sequences when the temperature is brought down, and in the next step 3) the primers are extended by the DNA polymerase enzyme by using the dNTPs.
After running the PCR reaction, you can "purify" the DNA products from all the other stuff present in the tube (by using special kits containing resins and buffers that will specifically bind to the DNA and then elute it) and, after checking that the product is the correct size (by "running a gel" with size markers next to the PCR product) analyze it further.
NUMEROUS (and ever-growing!) APPLICATIONS
By playing with some of the ingredients and the conditions of the PCR (mainly temperatures, number of cycles and primer sequences) one can adapt the technique for specific purposes. Initially, it was used to faithfully obtain multiple copies of a specific fragment of DNA to then get it sequenced or use it for cloning into a "plasmid" or "vector" or to introduce into an organism of choice like bacteria, yeast or mice.
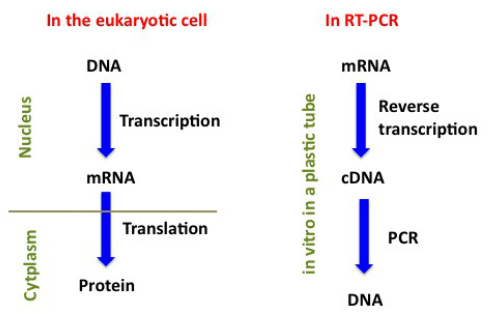
PCR can also be used to quantify gene expression when it is adapted to measure RNA levels - in this case, messenger RNA (mRNA) which is what the DNA in our genes is "transcribed" into. The mRNA, once made, travels from the nucleus of the eukaryotic cell into its cytoplasm, crossing the nuclear membrane, and once in the cytoplasm it is "translated' into its protein product which will perform a specific function, for which it might have to travel again, for example, into the nucleus if it happens to be a histone protein. If you have been following this far (some of this was on my "home" page) now you might get a bit confused by what happens in what's called "reverse transcriptase" PCR (RT-PCR). A comparison between what happens inside eukaryotic cells and RT-PCR is shown in the figure below. For RT-PCR, instead of DNA it is the mRNA which is extracted from the animal or plant sample obtained. Afterwards, an "RT" reaction is setup by which all the mRNA molecules present in the sample are "reverse transcribed" into complementary DNA (cDNA) once. This is made possible by using an enzyme that copies the single stranded mRNA into a complementary single stranded DNA and that we do not have in our cells but it is isolated from RNA viruses which use it to copy their RNAs into cDNAs. After the cDNA is made in the RT reaction, we can proceed with PCR to amplify the target sequence.
EXAMPLES OF MEDICAL AND PUBLIC HEALTH DIAGNOSIS THAT RELY MORE ON PCR
Most infectious agents detection methods in the past were developed to detect antibodies present in the blood of the infected individual, or alternatively infection can be detected by looking at blood samples under the microscope to see the organism directly using specific staining methods that facilitate visualization. Because the immune response to infection can take a few weeks, antibody (serological) tests can result in negative results when the infection is at an early stage. PCR (and other molecular assays which I haven't mentioned here) diagnosis, also from blood samples (extracted DNA or RNA) of the same infections can detect the causative agents much earlier. However, there is always a difference in the cost associated with molecular assays such as PCR which make them not as available as cheaper detection methods. In the field, especially in tropical areas, rapid diagnostic tests (RDTs) are usually based on detection of antibodies from the blood of the patient suspected to be sick when presenting with symptoms associated with malaria, HIV, tuberculosis, etc.
HIV diagnosis in infants: Some infections that are easily and cheaply detected by antibody tests, like HIV, have moved on towards detecting the viral DNA or RNA directly using PCR. For babies in Africa for example, where HIV incidence is quite high and many pregnant women are HIV-infected, antibody tests cannot provide a definitive diagnosis of HIV infection in their babies because maternal antibodies cross the placenta into the fetal circulation during pregnancy and remain detectable for more than a year after birth. Therefore the test will detect maternal antibodies and could result in a false positive HIV result when the baby might not have the virus, which would then translate into unnecessary treatment. PCR tests can now be done using DNA that comes from a dried blood spot from a filter paper obtained in remote rural areas such as African or Asian villages, which can be transported to a laboratory without a requirement for refrigeration. Once in the lab, PCR diagnostics is performed and the results are sent back to the village.
HPV diagnosis linked to risk of cervical cancer: HPV is the acronym for human papillomavirus, a group of more than 100 different related viruses that are spread during sexual contact in a skin-to-skin manner. During the annual PAP smear women should get at their gynecologist visits in developing countries, the sample extracted, consisting of cervical cells, is examined under the microscope by an expert cytologist. If the cells seen look "abnormal" in morphology (shape) then the next step is to do molecular diagnosis with specific primers to the known HPVs associated with high risk of developing cervical cancer. On a side note, a Gardasil vaccine is now available against HPV for females aged 9 to 26, approved by the FDA in 2006 consisting of a series of 3 shots over a 6-month period against 4 common types of HPV: 6, 11, 16, and 18. Types 6 and 11 or "high risk" are known to cause around 90% of genital warts cases, and types 16 and 18 are associated with up to 70% of cervical cancer cases. This vaccine is now also available for the prevention of genital warts in males 9 to 26 years of age. A new Cervarix HPV vaccine was approved by the FDA in 2009 that only prevents against HPV 16 and 18.
Risk for specific cancer types: For people with a known family history of certain cancers such as breast or ovarian cancer, or colon cancer, there are also special molecular (PCR-based) tests that can detect whether or not an individual in the family is at risk of developing the cancer given the presence of mutation (s) associated with this risk. For breast and ovarian cancer, these are mutations in the BRCA1 and BRCA2 genes.
Prenatal screening: From a sample containing fetal cells (chorionic villus, which can be obtained after 10 weeks of pregnancy, or amniocentesis after 15 weeks) many genetic conditions can be detected in the fetus, mainly trisomy 21 (Down syndrome) and trisomys 18 and 13- these are evident by the presence of 3 instead of two chromosomes for each of these pairs (a human karyptype figure showing all chromosome pairs is on my home page). As explained in my home page, these are routinely detected by experts using the microscope on samples of cultured amniotic cells, a process that takes weeks. There are PCR assays available to detect the same abnormalities that require just a few hr to process a sample, resulting in quick results and less anxiety for mothers with risk factors such as family history, advanced maternal age etc.

 RSS Feed
RSS Feed