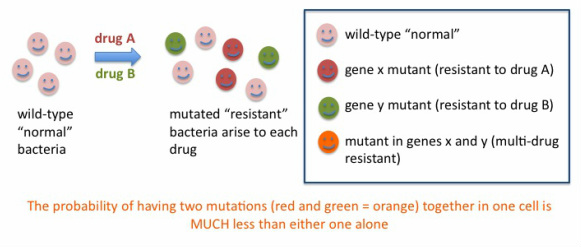
The most commonly heard of drug-resistant infection is probably MRSA: staph infections caused by methicillin-resistant Staphylococcus aureus. Staph infection was routinely treated in the 1950s with the antibiotic penicillin (check my home page for the amazing story of the fortuitous discovery of penicillin) which soon resulted in resistance of the bacterium to penicillin, and methicillin was developed as a chemically modified version of penicillin to counteract this resistance. Historically, many initially successful antibiotics have resulted in resistance arising in the target organisms, leading to the need for new antibiotics. A successful strategy to counteract the emergence of drug-resistant infection is the treatment with MORE THAN ONE antibiotic (a "cocktail") instead of just one ("monotherapy"). Below, the reason why this approach works is explained.
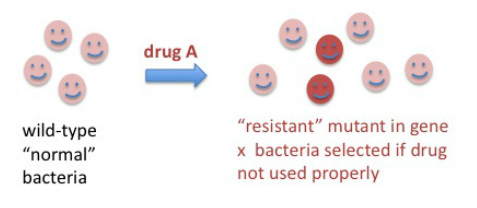
How do these drug-resistant organisms arise? These events happen by discrete mutations that occur spontaneously once in a while as cells divide and the enzymes responsible for DNA replication make mistakes. There are protective mechanisms inside cells that repair these mistakes in future DNA replication or repair events, therefore most mutations don't survive more than one or a few generations. However, when a mutation (or more than one mutation) occur that make cells "fitter" in the new environment, the mutation is "selected" by environmental pressure. In this case, the new environment is the presence of a drug. We have to keep in mind that most bacteria that cause infections divide rapidly inside our bodies, with many generations and cell divisions happening in one day.
One can calculate the probability of a mutation arising that would result in resistance to a particular drug, this probability is very, VERY small, in the range of 1 in 10 to the 6th to 1 in 10 to the 9th cell divisions. Still, if the drug is used in many people unnecessarily, and especially if not used as instructed but in shorter regimens or lower than recommended doses due to non-compliance or manufacturing issues, there is a population of bacteria that is not killed and might contain a proportion of drug-resistant bacteria. With time, the infection may recur and this time the resistant cells, if present, will take over quickly and then it will be hard to control them with the same antibiotic.
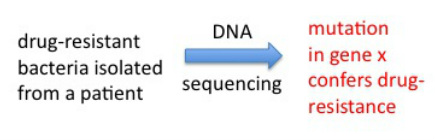
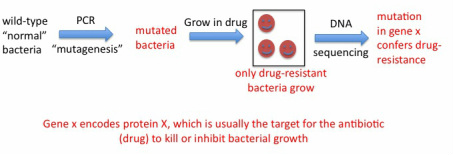
Mycobacterium tuberculosis, the bacterium that causes TB in humans, is not easy to work with in the lab due to a number of reasons. First, it grows extremely slowly, taking about one day to replicate once, which is the same doubling time as for mammalian (our) cells. So while you can grow many bacteria cultures both in the lab or in the clinic (for example, to test for drug-resistance of the patients' isolates to decide which drugs to treat with) in 24-48 hr, TB cultures take about 1 week minimum to grow. In addition, in the US and other developed countries, because TB is an airborne infection, it requires the use of a biosafety level 3 facility (BSL3). In the BSL3 controlled-access lab we wore respirator masks and a suit over our clothes, double layer of gloves and sterilized all materials coming out of the BSL3 every time we handled TB. BSL3 facilities are engineered so the flow of air is "negative" meaning nothing goes out, everything stays in with no recirculation. For these reasons, a non-pathogenic and much faster growing related mycobacteria is used sometimes as a "model" to work with to study TB in a faster, easier and non-hazardous manner. I used this bacteria, Mycobacterium smegmatis, to artificially generate mutants that were resistant to rifampicin and see which genes were altered in these mutants. The tool one can use to make these mutants fast is PCR. PCR is normally used to faithfully "amplify" DNA from very few copies (see details in my homepage). However, one can play with certain conditions of the assay (mostly temperature) to have the polymerase enzyme that replicates the DNA in the assay make mistakes at a high rate. This is called "in vitro mutagenesis". You can take a piece of DNA - gene of interest - and using these conditions generate a pool of mutated copies of your gene, then "integrate" these into cultures of your bacteria that you want to mutagenize. Afterwards, you grow the bacteria in the presence of rifampicin, and only the resistant mutants should grow. We did this, and obtained several rifampicin mutants that we sequenced to show one or more mutations in the rpoB gene, which confirmed this gene as the molecular basis for rifampicin resistance in mycobacteria.

 RSS Feed
RSS Feed